Comprehensive List of Researchers "Information Knowledge"
Department of Complex Systems Science
- Name
- AOKI, Setsuyuki
- Group
- Life-Science Informatics Group
- Title
- Associate Professor
- Degree
- Dr. of Science
- Research Field
- Biological rhythms / Gene network / Molecular biology

Current Research
Generation of Biological Functions by the Interactions among Information Bio-molecules
■Outline■Living systems display a wide variety of cyclic events. Circadian rhythms with an endogenous periodicity of about 24 hours are particularly ubiquitous phenomena. Recent studies on many model organisms have shown that circadian clocks, the core systems generating these rhythms, are composed of regulatory networks among many"clock genes"and their protein products"clock proteins". A circadian clock, therefore, can be an excellent model for studying biological functions generated from the interaction among information bio-molecules. We are studying the molecular mechanisms of the clock by using the moss Physcomitrella patens, which is suitable for our purpose because gene targeting techniques are applicable to this primitive plant. It is becoming possible to understand the evolution of biological systems in light of genome dynamics because various genome and post-genome projects are in progress. Regarding plants, circadian clocks have been intensively studied in plants such as rice and arabidopsis. P. patens diverged from those "higher" plants at a very early stage of the evolution of land plants. The genome project on P. patens, started in 2004, is entering its final stages ; the first draft sequence will soon be available, and its annotation process has already started. It will be possible to understand the evolution of circadian clocks as changes of the network structures of clock genes and clock proteins, through studying the clock mechanism of P. patens and comparing genomic information from various organisms.
■TOPCS■
(1) Identification and analysis of the regulatory network of clock genes in P. patens The mechanisms of the clocks in eukaryotes, including plants, are supposed to be based on negative feedback loops in the regulatory networks of clock genes. Clock genes such as CCA1, LHY, PRR1(TOC1) and PCL1 have been cloned in the model plant arabidopsis ; Alabadi et al. (2001) proposed a model which is based on the transcriptional negative feedback loop comprising the former three clock genes. We have identified candidate clock genes from P. patens, which are orthologous to the arabidopsis clock genes. An aim of this research is to unveil the expression dynamics and interactions of these genes, and to conduct functional analyses of these genes by gene targeting techniques. Based on such an approach, we will endeavor to understand the molecular mechanism of the circadian clock. Furthermore, we will investigate the evolution of circadian clocks, which are most probably based on diversification and multiplication of clock genes and changes of the network structures of these genes.
(2) The mechanisms by which timing information is transmitted from the nucleus to chloroplasts. Chloroplasts are semi-autonomous organellae derived from endosymbiosis of a photosynthetic bacterium. Therefore, chloroplasts have their own genomes, which have 100 to 200 genes. Although some of these chloroplast genes are known to express circadian rhythms in their expression, their regulatory mechanisms are almost totally unknown. Sigma, the nuclear-encoded RNA polymerase subunit, is likely candidate for a critical factor in circadian regulation on the chloroplast gene expression. We want to understand the molecular mechanisms conveying timing information from the nuclear clock to the chloroplast genome by examining the functions of sigma genes in P. patens.
■Future work■
We aim to comprehensively understand the molecular mechanisms and evolution of circadian clock by using "Omics" related information. We will also try to establish other effective experimental systems that make it possible to understand the generation and evolution of biological functions from a more general point of view.
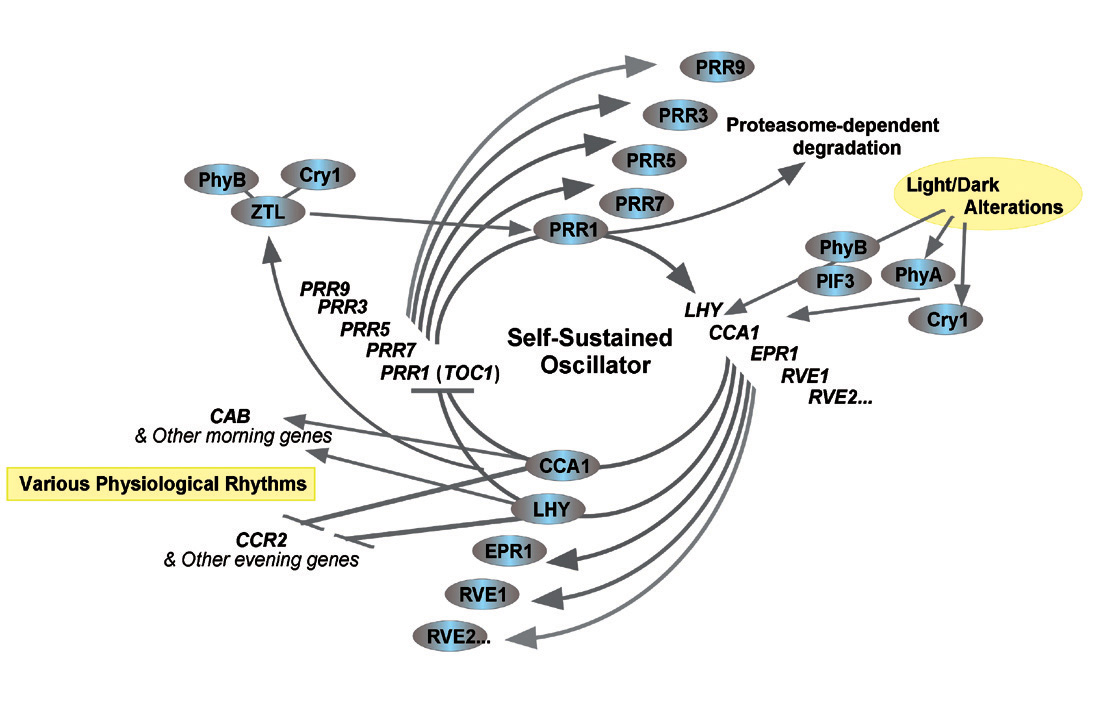
Figure : Model of the circadian clock in plants
Career
- Setsuyuki Aoki received the Dr. of Science degree from the Graduate School of Science, Kyoto University in 1996.
- He was a research associate from 1996 to 2003 and an associate professor from 2003 at the Graduate School of Human Informatics, Nagoya University, and since 2003, he has been an assistant professor at the Graduate School of Information Science, Nagoya University.
Academic Societies
- Japanese Society for Chronobiology
- The Genetics Society of Japan
- The Japanese Society of Plant Physiologists
- The Botanical Society of Japan
Publications
- Pseudo-Response Regulator (PRR) homologues of the moss Physcomitrella patens: Insights into the evolution of the PRR Family in land plants. DNA Research 18(1):39-52 (2011)
- Heterologous expression and functional characterization of a Physcomitrella Pseudo Response Regulator homolog, PpPRR2, in Arabidopsis. Bioscience, Biotechnology, and Biochemistry 75(4):786-789 (2011)
- Functional characterization of CCA1/LHY homolog genes, PpCCA1a and PpCCA1b, in the moss Physcomitrella patens. Plant Journal 60(3):551-563 (2009)








